Contents
 Basic Translation Process
Basic Translation Process
 The Genetic Code
The Genetic Code
 Reading frame of a sequence
Reading frame of a sequence
 Start/stop codons
Start/stop codons
 Degeneracy of the genetic code
Degeneracy of the genetic code
 Origin of the genetic code
Origin of the genetic code
 Reverse Translation
Reverse Translation
A particular protein follows from the translation of a DNA sequence whereas the reverse translation needs not have a specific solution according to the Genetic Code. Because of the degeneracy of Genetic Code one amino acid may be coded by alternative codons. This degeneracy leads to ambiguity in back translation process. So in this codon translation program codon are restricted for the back translation and one can choose user define restricted codons or from tehe extremophilics microorganisms codon usage. Translation is a process where genetic information is translated from a 'nucleic acid language' to an 'amino acid language'. Translation is catalyzed by a large enzyme called a ribosome, which contains proteins and ribosomal RNA (rRNA). Translation also involves specific RNA molecules called transfer RNA (t-RNA) which can bind to three basepair codons on a messenger RNA (mRNA) and also carry the appropriate amino acid encoded by the codon. The ribosome assembles on the first AUG (start codon) in the mRNA. This codon encodes the amino acid methionine (Met).
The Genetic Code
In 1968 the Nobel Prize in Medicine was awarded to Robert W. Holley, Har Gobind Khorana and Marshall W. Nirenberg for their
interpretation of the Genetic Code (http://nobelprize.org/medicine/laureates/1968). The genetic code is the set of rules by which
information encoded in genetic material (DNA or RNA sequences) is translated into proteins (amino acid sequences) by living
cells. The code defines a mapping between tri-nucleotide sequences, called codons, and amino acids. We are now using computer
programs to translate DNA sequences into predicted amino acid sequences.
The code in which genetic instructions are written, using an alphabet based on the four bases in DNA and RNA: adenine, cytosine, guanine, and thymine (for DNA) or uracil (for RNA). Each triplet of bases indicates that a particular kind of amino acid is to be synthesized. Since there are 20 amino acids and 64 possible triplets, more than one triplet can code for a particular amino acid. The code is non-overlapping; the triplets are read end-to-end in sequence (eg UUU = phenylalanine, UUA = leucine, CCU = proline); and there are three triplets not translated into amino acid, indicating chain termination.
The code is universal and applies to all species. Theoretically, there are 4³ = 64 different codon combinations possible with a triplet codon of three nucleotides. In reality, all 64 codons of the standard genetic code are assigned for either amino acids or stop signals during translation. If, for example, an RNA sequence, UUUAAACCC is considered and the reading-frame starts with the first U (by convention, 5' to 3'), there are three codons, namely, UUU, AAA and CCC, each of which specifies one amino acid. Table 2 shows what codons specify each of the 20 standard amino acids involved in translation.
[TOP]
Table 1: RNA codon table
This table shows the 64 codons and the amino acid each codon codes for. (see the full names of the amino acids
here.)
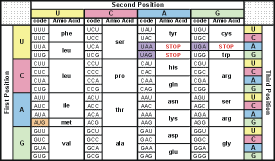
Table 2: Synonymous Codon for Back Translation
A particular protein follows from the translation of a DNA sequence whereas the reverse translation needs not have a specific solution according to the Genetic Code. Because of the degeneracy of Genetic Code one amino acid may be coded by alternative codons. This degeneracy leads to ambiguity in back translation process. So in this codon translation program codon are restricted for the back translation and one can choose user define restricted codons or from tehe extremophilics microorganisms codon usage.
| Ala/A | GCU, GCC, GCA, GCG | Leu/L | UUA, UUG, CUU, CUC, CUA, CUG |
|---|---|---|---|
| Arg/R | CGU, CGC, CGA, CGG, AGA, AGG | Lys/K | AAA, AAG |
| Asn/N | AAU, AAC | Met/M | AUG |
| Asp/D | GAU, GAC | Phe/F | UUU, UUC |
| Cys/C | UGU, UGC | Pro/P | CCU, CCC, CCA, CCG |
| Gln/Q | CAA, CAG | Ser/S | UCU, UCC, UCA, UCG, AGU, AGC |
| Glu/E | GAA, GAG | Thr/T | ACU, ACC, ACA, ACG |
| Gly/G | GGU, GGC, GGA, GGG | Trp/W | UGG |
| His/H | CAU, CAC | Tyr/Y | UAU, UAC |
| Ile/I | AUU, AUC, AUA | Val/V | GUU, GUC, GUA, GUG |
| START | AUG | STOP | UAG, UGA, UAA |
[TOP]
Reading frame of a sequence:-
A codon is defined by the inital nucleotide from which translation starts. For example, the string GGGAAACCC, if read from
the first position, contains the codons GGG, AAA and CCC;and if read from the second position, it contains the codons GGA
and AAC;
The actual frame a protein sequence is translated in is defined by a start codon, usually the first AUG codon in the mRNA
sequence.
Start/stop codons:-
Translation starts with a chain initiation codon (start codon). The most common start codon is AUG, which also codes for
methionine, but other start codons are also used. Stop codons are also called termination codons and they signal release
of the nascent polypeptide from the ribosome due to binding of release factors in the absence of cognate tRNAs with
anticodons complementary to these stop signals.
Degeneracy of the genetic code:-
Many codons are redundant, meaning that two or more codons can code for the same amino acid.
Degeneracy results because a triplet code designates 20 amino acids and a stop codon. For example, if there were two bases
per codon, then only 16 amino acids could be coded for (42=16). For example, in theory, four-fold degenerate codons can
tolerate any point mutation at the third position, although codon usage bias restricts this in practice in many organisms;
A practical consequence of redundancy is that some errors in the genetic code only cause a silent mutation or an error
that would not affect the protein because the hydrophilicity or hydrophobicity is maintained by equivalent substitution
of amino acids; for example, a codon of NUN (where N = any nucleotide) tends to code for hydrophobic amino acids.
These variable codes for amino acids are allowed because of modified bases in the first base of the anticodon of the tRNA,
and the base-pair formed is called a wobble base pair.
In certain proteins, non-standard amino acids are substituted for standard stop codons, depending upon associated signal
sequences in the messenger RNA: UGA can code for selenocysteine and UAG can code for pyrrolysine as discussed in the relevant
articles.
Origin of the genetic code:-
Despite the variations that exist, the genetic codes used by all known forms of life on Earth are very similar. Although
much circumstantial evidence has been found to indicate that originally the number of different amino acids used may have
been considerably smaller than today, precise and detailed hypotheses about exactly which amino acids entered the code in
exactly what order has proved far more controversial. A third is that natural selection organized the codon assignments of
the genetic code to minimize the effects of genetic errors (mutations).
[TOP]
Reverse Translation
In all living cells which contain hereditary material such as DNA, a transcription to mRNA and subsequent a translation to
proteins occur. This is of course simplified but is in general what is happening in order to have a steady production of
proteins needed for survival of the cell. In bioinformatics analysis of proteins it is sometimes useful to know the
ancestral DNA sequence in order to find the genomic localization of the gene. Thus, the translation of proteins back to
DNA/RNA is of particular interest, and is called reverse translation or back-translation.
The Genetic Code represents translations of all 64 different codons into 20 different amino acids. Therefore it is no
problem to translate a DNA/RNA sequence into a specific protein. But due to the degeneracy of the genetic code, several
codons may code for only one specific amino acid.
Challenge of reverse translation
A particular protein follows from the translation of a DNA sequence whereas the reverse translation needs not have a
specific solution according to the Genetic Code. The Genetic Code is degenerate which means that a particular amino acid
can be translated into more than one codon. Hence there are ambiguities of the reverse translation.
Solving the ambiguities of reverse translation
In order to solve these ambiguities of reverse translation you can define how to prioritize the codon selection, e.g:
* Choose a codon randomly.
* Select the most frequent codon in a given organism.
* Randomize a codon, but with respect to its frequency in the organism.
As an example we want to translate an alanine to the corresponding codon. Four different codons can be used for this
reverse translation; GCU, GCC, GCA or GCG. By picking either one by random choice we will get an alanine.
The most frequent codon, coding for an alanine in E. coli is GCG, encoding 33.7% of all alanines. Then comes GCC (25.5%),
GCA (20.3%) and finally GCU (15.3%).
By selecting codons from a distribution of calculated codon frequencies, the DNA sequence obtained after the reverse
translation, holds the correct (or nearly correct) codon distribution. It should be kept in mind that the obtained DNA
sequence not necessarily is identical to the original one encoding the protein in the first place, due to the degeneracy
of the genetic code.
In order to obtain the best possible result of the reverse translation, one should use the codon frequency table from the
correct organism or a closely related species. In this program we are taking the codons of highest frequency from the codon
usage table of Escherichia coli.
[TOP]

